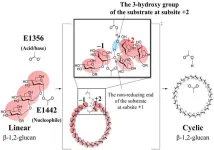
The polysaccharide β-1,2-glucan consists of repeating units of glucose linked together by β-1,2-glycosidic bonds. Cyclic β-1,2-glucans (CβGs) occur in different bacterial species and have a role in bacterial infections and symbiotic relationships. CβG biosynthesis is catalyzed by cyclic β-1,2-glucan synthase (CGS), an enzyme that catalyzes the cyclization (closed ring formation) of linear β-1,2-glucan (LβG).
Since the method for large-scale enzymatic synthesis of linear β-1,2-glucan has already been established, combining it with this enzyme is technically feasible for efficient one-pot synthesis of cyclic β-1,2-glucan. However, the cyclizing activity of this enzyme is not very strong, and it also has low stability as an enzyme.
Previous studies have revealed that the bonding of glucose to CGS, the elongation of LβG chains, glucan chain length adjustment, and cyclization of the glucans are spread across three distinct domains of CGS. However, while it is known that the enzyme domain in the middle region of CGS cyclizes LβGs, a detailed mechanism for the process has eluded scientists until now.
Recently, a team of researchers from Japan has uncovered the catalytic mechanism of CGS cyclization following functional and structural analyses of the CGS cyclization domain from the bacteria Thermoanaerobacter italicus (TiCGSCy). The team was led by Assistant Professor Nobukiyo Tanaka from the Department of Applied Biological Science at Tokyo University of Science (TUS) and included Br. Ryotaro Saito, Associate Professor Masahiro Nakajima, and Associate Professor Tomoko Masaike, all from TUS, as well as Associate Professor Hiroyuki Nakai from Niigata University and Dr. Kaito Kobayashi from the National Institute of Advanced Industrial Science and Technology (AIST).
Their findings have been published in the journal Applied Microbiology and Biotechnology on 1 February 2024. Dr. Tanaka elaborates, “CGSs are important enzymes from a physiological standpoint as CβGs are implicated in various bacterial diseases and symbiotic relationships. We were keen to provide insights into the structure and function of CGSs, which are receiving attention in research but for which the mechanism of β-1,2-glucan cyclization remains a mystery.”
The first step of the research involved expressing the cyclization domain alone, TiCGSCy, as a recombinant enzyme in Escherichia coli. The next step toward characterization involved analyzing reaction products when LβGs were used as a substrate for TiCGSCy. “We discovered that TiCGSCy produced β-glucosidase-resistant compounds. On examining them using nuclear magnetic resonance spectroscopy, we found CβGs. This was the first evidence that TiCGSCy produced CβGs,” explains Dr. Tanaka.
To determine the action patterns of TiCGSCy on LβGs , the team treated TiCGSCy with β-1,2-gluoligosaccharides (LβGs that comprise 2–10 glucose units) and analyzed the products. The reaction products revealed that the TiCGSCy mechanism lacked a hydrolysis reaction, featured transglycosylation, and required substrates that were at least hexasaccharide (polymers of six or more glucose units) in length. This finding matched data from previous studies where related CGSs produced CβGs with polymers of around 20 glucose units. Finally, it has been suggested that TiCGSCy has an anomer-retaining mechanism, as it generates a product with the same anomer as its substrate.
Further, structural analysis of TiCGSCy using X-ray crystallography showed that TiCGSCy, the β-1,2-glucanases from Chitinophaga pinensis (CpSGL, belonging to GH144) and the fungus Talaromyces funiculosus (TfSGL, belonging to GH162) are structurally similar, though the amino acid sequence homology of them is very low.
The complex structure of CpSGL (GH144), the Michaelis complex structure of TfSGL (GH162), and the overall structure of TiCGSCy were superimposed to explore the catalytic residues of TiCGSCy. As a result, E1356 was found to be located in a position to act on the oxygen atom of the glycosidic bond at the cleavage site via the 3-OH group of the glucose molecule at subsite +2, and E1442 was positioned to directly perform a nucleophilic attack on the anomeric center of subsite –1. Consequently, they were inferred to be the general acid/base and the nucleophilic residue for catalysis, respectively.
The detailed reaction mechanism of TiCGSCy is as follows:
(a) E1356 acts as a general acid for catalysis, donating a proton to the oxygen atom via the 3- OH group of the glucose molecule at subsite +2. Simultaneously, E1442 acts as a nucleophile, attacking the anomeric center at subsite –1 to form a glycosyl-enzyme intermediate.
(b) E1356 acts as a general base, drawing a proton from the 2-OH group of the glucose molecule at subsite +1 via the 3-OH group of the glucose molecule at subsite +2.
(c) The activated 2-OH group at subsite +1 attacks the anomeric carbon at subsite –1, releasing the glycosyl transfer product.
In the reaction mentioned above, it was suggested that when the hydroxyl group from a molecule different from the one forming the glycosyl-enzyme intermediate attacks the anomeric carbon atom in (a), a linear product is obtained. Conversely, when the attacking hydroxyl group is from the same molecule (Fig. 1), a cyclic product is formed.
Overall, these findings have significant implications for the characterization of TiCGSCy. “Given its noncanonical reaction mechanism, this CGS defines a new family of glycoside hydrolases, GH189,” says Dr. Tanaka.
In conclusion, through this research, the researchers have identified residues important for cyclizing activity, which leads to the desired search for enzymes with stronger cyclizing activity and higher stability. The team is confident that their work will open research avenues that explore the inhibition of CGS.
***
Reference
Title of original paper: Functional and structural analysis of a cyclization domain in a cyclic β-1,2-glucan synthase
Journal: Applied Microbiology and Biotechnology
DOI: https://doi.org/10.1007/s00253-024-13013-9
Authors: Nobukiyo Tanaka1, Ryotaro Saito1, Kaito Kobayashi2, Hiroyuki Nakai3, Shogo Kamo1, Kouji Kuramochi1, Hayao Taguchi1, Masahiro Nakajima1 and Tomoko Masaike1
Affiliations:
1Department of Applied Biological Science, Faculty of Science and Technology, Tokyo University of Science
2Artificial Intelligence Research Center, National Institute of Advanced Industrial Science and Technology (AIST)
3Faculty of Agriculture, Niigata University
Further Information
Assistant Professor Nobukiyo Tanaka
Department of Applied Biological Science, Faculty of Science and Technology
Tokyo University of Science
Email: n_tanaka@rs.tus.ac.jp
Associate Professor Masahiro Nakajima
Department of Applied Biological Science, Faculty of Science and Technology
Tokyo University of Science
Email: m-nakajima@rs.tus.ac.jp
Associate Professor Tomoko Masaike
Department of Applied Biological Science, Faculty of Science and Technology
Tokyo University of Science
Email: tmasaike@rs.tus.ac.jp
Associate Professor Hiroyuki Nakai
Faculty of Agriculture
Niigata University
Email: nakai@agr.niigata-u.ac.jp
Media contact
Hiroshi Matsuda
Public Relations Divisions
Tokyo University of Science
Email: mediaoffice@admin.tus.ac.jp
Website: https://www.tus.ac.jp/en/mediarelations/
Public Relations office
Niigata University
Email: pr-office@adm.niigata-u.ac.jp
Funding information
This work was supported by Photon Factory for X-ray data collection (Proposal No. 2020G527 and No. 2021G685).
END