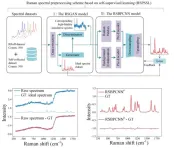
Raman spectral preprocessing
Raman spectroscopy detects the molecular bond information of the chemical components of the sample in situ in a non-destructive and label-free manner (Terms >>>). It is an emerging metabolic-related spectromics technology in biological and clinical medical research and is expected to promote the evolution of precision medicine. At present, Raman spectroscopy is applied in biomedical detections of body fluids, exosomes, cells/microbes, and tissues, among which Raman hyperspectral imaging of biological tissues can provide three-dimensional spatial distribution information of chemical molecular bonds and is expected to be a powerful tool for the screening and research of molecular mechanisms of physical and disease occurrence and development.
However, it is susceptible to interference from instruments, environmental noise, and background signals (non-Raman signals/baselines) due to weak Raman scattering signals. The superposition of these noises and baseline signals seriously affects the separation and resolution of the Raman spectroscopy intrinsic signals (Raman peaks), which limits the application and popularization of Raman spectroscopy, especially across instruments, samples, and spectral types. The complexity of the components of biological samples and the severe interference of fluorescence signals makes the biomedical application of Raman spectroscopy difficult. Spectral preprocessing methods with efficient spectral noise removal and high-fidelity baseline correction capabilities are the prerequisites and challenges for high-quality Raman spectroscopy applications.
Challenges
Traditional numerical analysis methods require multiple manual parameter adjustments to achieve better noise reduction and baseline correction effects unsuitable for high-throughput application scenarios such as clinical diagnosis and hyperspectral image processing.
Existing deep learning preprocessing methods have the advantage of auto-parameter tuning. However, the noise removal and baseline correction spectral fidelity still need to be improved to achieve universal application across instruments and samples.
The performance of conventional supervised deep learning spectral preprocessing methods relies on high-quality label data training. This kind of label data is free of noise and baselines, where noise can be reduced by averaging multiple acquisition spectra, while the baselines are hard to eliminate by conventional instrument acquisition. In addition, the deep learning model trained by this method needs to re-collect data and retrain when used across instruments and samples. Their generalization ability needs to be improved.
When the mathematical simulation dataset is used for training, although it can be adapted across instruments and samples, due to the difference between the actual spectral noise/baseline and the numerical simulation, the spectral fidelity of the preprocessing model is still insufficient for biomedical sample Raman hyperspectral images preprocessing.
Raman spectral preprocessing algorithm with high fidelity and generalization ability
Recently, the team of Professor Perry Ping Shum from the Department of EEE, Southern University of Science and Technology, State Key Laboratory of Optical Fiber and Cable Manufacture Technology, Guangdong Key Laboratory of Integrated Optoelectronics and Intellisense, and their collaborators proposed a two-step Raman spectral preprocessing strategy (RSPSSL) based on self-supervised learning to achieve high-fidelity spectral denoising and baseline correction across instruments, samples and spectral types, and promote the chemical resolution visualization of Raman hyperspectral images of clinical tissue samples.
The article "RSPSSL: A Novel High-fidelity Raman Spectral Preprocessing Scheme to Enhance Biomedical Applications and Chemical Resolution Visualization" was published in Light: Science & Applications. Ph.D. candidate Jiaqi Hu is the first author, and Associate Researcher Gina Jinna Chen (co-first author) and Professor Perry Ping Shum are the corresponding authors.
The first step of the scheme is to establish a self-supervised model according to the mutual independence of the physical relationship between Raman peaks, noise, and baselines, self-decompose, rearrange and reconstruct the unlabeled training spectra, and build a generative adversarial network to obtain the ability to generate an infinite number of labeled high-simulation Raman spectral pairs, to solve the problem of unlabeled real Raman spectra.
The label-free training spectra employ diverse data from multiple laboratories across instruments, samples, and spectral types to obtain the diversity of noise and baselines. Secondly, to adapt to the complexity of actual spectral data, the preprocessing model enhances the fitting ability of complex signals through the end-to-end connection of multiple submodules. The preprocessing model RSBPCNN# can be used for Raman spectral preprocessing from any instrument, sample, and spectral type without manual intervention or retraining.
The preprocessing model RSBPCNN# has excellent noise removal and baseline correction capabilities, and the processed spectral fidelity is high. This ability to extract weak signals with different signal-to-noise ratios reduces sampling time and improves downstream applications.
Versatile Raman spectral preprocessing
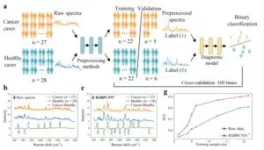
We used multiple experiments to verify the model's generalizability in this study. Without any changes, the preprocessing model RSBPCNN# was applied directly to cancer diagnosis, herbicide concentration prediction, and hyperspectral imaging, and it significantly improved the application effects. This method significantly improves the diagnostic and concentration prediction accuracy in the case of few-shot applications and further verifies the fidelity of spectral preprocessing. At the same time, these experimental data come from different instruments and laboratories, demonstrating their cross-instrument adaptability.
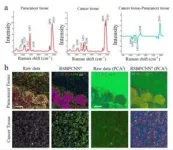
Hyperspectral image quality improvement
The most unique capability of Raman hyperspectral images is volumetric chemical imaging. However, the weak bioimaging signal is not visible due to the superimposed baseline signals. Applying the preprocessing model RSBPCNN#, the Raman peak intensities were restored to reconstruct chemically specific images. At the same time, this method can also significantly improve the signal-to-noise ratio and reduce the sampling time by dozens of times.
Summary and outlook
In this study, RSPSSL, a new strategy for self-supervised two-step Raman spectral preprocessing, is proposed to obtain and generate an infinite number of labeled high-fidelity simulation spectral datasets through the fine separation and reconstruction of diverse spectral features through exquisite algorithm model design and to train and optimize the predominant preprocessing model with high fitting ability to obtain the purpose RSBPCNN# with high robustness. The model can achieve a high throughput of arbitrary Raman spectroscopy noise elimination and baseline correction without human intervention. Because of its spectral high-fidelity characteristics, it can significantly improve the accuracy of cancer diagnosis and solution concentration prediction in experiments, improve the full-spectrum quality of full hyperspectral images, eliminate the background signal of the biological silent zone, realize the visualization of the chemical resolution of images in the spectral fingerprint region, and reflect the broad-spectrum applicability across instruments, samples, and spectral types. In the future, the resolution of hyperspectral images can be further improved by involving the spatial distribution of spectra.
Software Sharing
This method has been integrated into the laboratory-sharing platform for scientific use. Researchers can load Raman spectral data in batches to achieve rapid spectral preprocessing (1900 spectra/sec). URL links: https://github.com/oilab-sustech/RSPSSL .
END