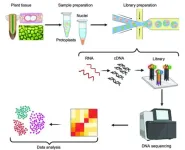
Recent breakthroughs in single-cell RNA sequencing (scRNA), such as the recently developed “RevGel-seq” method, has revolutionized plant cell analysis. This technique, independent of special instruments, streamlines processes and resolves protoplast isolation challenges. Now, a multinational team of researchers review this and other such recent advances in plant scRNA sequencing with the intention of providing guidance for facilitating the appropriate selection of scRNA methods for different plant samples.
In the world of plant biology, understanding the intricacies of individual plant cells has been a complex challenge, particularly due to the unique structure of these cells encapsulated by rigid cell walls. This challenge has, in turn, hindered the isolation of intact nuclei or protoplasts, which are essential for in-depth analysis. Now, while traditional RNA sequencing has played an instrumental role in helping researchers understand the intricacies of plant cells, the breakthrough development of single-cell RNA (scRNA) sequencing has enabled a groundbreaking advancements in our understanding and analysis of plant biosystems.
Moreover, other technical advancements in scRNA sequencing, such as the development of protocols for cell isolation, library preparation, and sequencing technologies have furthered the ability to overcome plant-specific challenges in analysis. While studies in the past have reviewed the utility and practical applicability of scRNA sequencing, not many have attempted to look at the technological developments with regards to scRNA sequencing. Now, however, in a recent review published in BioDesign Research on 29 February, 2024, researchers have comprehensively looked at the recent advances and paradigm shifts in plant systems biology. Plant systems, with their complex cellular architecture, demand innovative approaches to unravel the secrets hidden within plant cells. The review captures these recent developments in scRNA technology, besides discussing the challenges and future potential opportunities in this field.
The review first discusses the methods popularly used in single-cell transcriptomics, which include experimental and computational components, and also elaborates on the general workflow for scRNA sequencing. Given the challenges and limitations of protoplast preparation and nuclei isolation methods, the use of microfluidics platform offers a primary workflow for single-cell isolation, separation, and analysis. Moreover, while the choice between the protoplast and nuclei for plant scRNA sequencing depends on the research objective, the review urges researchers to carefully consider the choice of multiple factors, such as research goal, plant species, and other trade-offs, while determining the most suitable method.
The review then touches upon the advancements in scRNA sequencing library construction and sequencing, highlighting the significant achievements, such as the Nextera XT DNA Library Preparation Kit, the BD Rhapsody system, and Illumina sequencing technologies. Next, the researchers discuss the multiple steps involved in scRNA sequencing, before moving on to discuss the recent developments in scRNA databases. PlantscRNAdb, for instance, was developed for analyzing scRNA-seq data in plants, and contains 26,326 marker genes. Plant Cell Marker DataBase, on the other hand, contains 81,117 cell marker genes of 263 cell types in 22 tissues across six plant species. They then also discuss the improvements made by researchers to address the limitations with plant scRNA databases, such as “scPlantDB,” a comprehensive database created by He et al., covering around 2.5 million cells across 17 plant species.
The researchers further go on to discuss the applications of scRNA sequencing in plant systems biology and elaborate how scRNA sequencing has been used to study plant biology at the cellular level, shedding light on molecular mechanisms of plant development, plant responses to biotic and abiotic stresses, epigenetic regulation in plants, cell fate determination and organogenesis, among others.
While discussing the challenges and potential for further development of scRNA sequencing, the researchers highlight a recent breakthrough development aimed at optimizing protoplast isolation methods and establishing standardized single-cell RNA sequencing (scRNA-seq) processes across diverse laboratories. Employing a multifaceted approach, the development sought to combined innovative scRNA-seq technologies with traditional methods to tackle the intricate world of plant cells. Sample preparation became a meticulous process, focusing on variables like enzyme treatment duration, temperature, and osmotic potential. Notably, the study explored novel scRNA-seq technologies, including RevGel-seq, originally developed for animal systems but now introduced to the realm of plant research.
Adding further, Dr. Xiaohan Yang, the lead researcher of this review affiliated with the Oak Ridge National Laboratory, explains, “RevGel-seq, a recently developed breakthrough in scRNA-seq methods, is truly a game-changer. Unlike traditional methods that rely on specific single-cell RNA instruments during sample preparation, RevGel-seq operates on cell-barcoded bead complexes. This innovation sample preparation for scRNA-seq in human and mouse cell not only convenient but also highly efficient.” RevGel-seq also eliminates the need for specific instruments, providing flexibility in sample collection and processing at different times or locations. Needless to say, the results surpassed expectations, showcasing RevGel-seq's potential to reshape how single-cell RNA sequencing is conducted in plant research.
The recent advancements enlisted in this review have significant implications for the future of plant biology. While scRNA sequencing has indeed emerged as a groundbreaking technology in the field of plant systems biology and synthetic biology, the review sheds light on the possible path for addressing existing challenges and gives directions for future researchers. It also highlights the need to integrate scRNA sequencing technologies with other omics technologies and with other computational tools for enhancing the understanding of complex cellular interactions in plants.
In conclusion, the review suggests a positive way forward in the field of plant biology, with single-cell RNA sequencing for plant cells shedding light on their inner workings and opening doors for deeper insights. Besides addressing current hurdles, it can serve as a guide for researchers towards a more sustainable future with notable breakthroughs.
###
References
Journal: BioDesign Research
Titles of original papers: Advances in the application of single-cell transcriptomics in plant systems and synthetic biology
DOI: 10.34133/bdr.0029
END