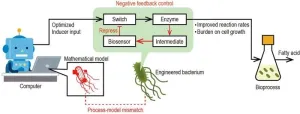
Control systems are ubiquitous in modern day technology. In industrial contexts, these systems ensure that relevant variables remain within a desirable range to keep processes running safely and efficiently. A vast array of control strategies exists, and it is not uncommon to combine different types of controllers to improve performance. For instance, high-level controllers based on mathematical modeling of a given process are routinely combined with low-level controllers, such as the widely used Proportional–Integral–Derivative (PID) controller, which does not rely on models and instead uses real-time feedback to adjust control actions.
Combining different controllers is not, however, a straightforward endeavor. In biosynthesis plants, where engineered bacteria are used to produce desired products, the concentrations of specific substances inside the microorganisms represent important control variables. Not only are these variables difficult to measure accurately but control systems must also address inaccuracies between the mathematical models used and the actual process. Addressing these process-model mismatches (PMMs) is a significant challenge in controlling biosynthetic processes.
Fortunately, a research team led by doctoral student Tomoki Ohkubo from the Graduate School of Science and Technology at Nara Institute of Science and Technology and Associate Professor Katsuyuki Kunida from Fujita Health University, Japan, has been actively investigating a promising solution to this problem. In their latest study published in volume 14 of Scientific Reports on November 18, 2024, they present a convincing proof-of-concept for a hybrid ‘in silico/in-cell’ controller (HISICC). The team’s technology combines a computer-driven, model-based optimization controller with a feedback control mechanism engineered directly into an Escherichia coli (E. coli) bacteria strain engineered for producing fatty acids. Mr. Tomoki Ohkubo from the Nara Institute of Science and Technology also contributed to the research and was the lead author of the study.
To demonstrate the potential of HISICC, the team compared three different control strategies for the E. coli-based synthesis of fatty acids via simulations. The first approach used a previously developed “FA2” strain, which has an engineered regulator that can drive up the production of the key “bottleneck” enzyme acetyl-CoA carboxylase (ACC) when induced by isopropyl-β-D-thiogalactopyranoside (IPTG). While the overexpression of ACC increases fatty acid yield, excessive strain on the bacteria can lead to cytotoxicity. As a result, this “no brakes” approach is the most rudimentary form of control technique.
In contrast, the HISICC approach uses a previously developed “FA3” strain, which contains an in-cell feedback loop. “In this design, if the ACC level becomes too high, increased levels of a responsive transcription factor trigger the production of a lactose repressor protein, which in turn represses ACC overexpression,” explains Mr. Ohkubo, “Meanwhile, like in FA2, IPTG can be administered externally to drive up ACC expression.” The third and final approach employed a newly designed strain called “FA4.” This bacterial strain did not have a feedback loop but instead contained a second “input channel” to drive up the production of lactose repressor protein. Simply put, FA4 has an externally controlled “brake system,” unlike FA2.
The team carefully developed mathematical models for each bacterial strain and conducted a series of simulations addressing different types of PMMs. As expected, the FA3 and FA4 strains performed better than FA2. This improved performance was attributed to the ability to tune down ACC overexpression before bacterial strains are negatively affected, meaning that enforcing a higher initial ACC expression was possible, leading to a higher initial yield. Notably, among the three methods tested, the HISICC approach (FA3) achieved the highest fatty acid yield. “Our findings demonstrate the potential of HISICC with intracellular biosensing as a solution to the PMM problem,” remarks Dr. Kunida. “Considering that the in-cell controller used in this study is rather simple in structure and crude in operation, we expect that a more advanced in-cell controller capable of differential or second-order differential control would reduce response times and markedly improve performance.”
Together, the techniques and results featured in this study could form the basis for new control strategies in complex industrial bioprocesses. By improving the production efficiency of fuels and important chemicals, the HISICC approach has the potential to reduce costs and minimize the environmental impact associated with these processes.
***
Reference
Title of original paper: A hybrid in silico/in-cell controller that handles process-model mismatches using intracellular biosensing
Journal: Scientific Reports
DOI: https://doi.org/10.1038/s41598-024-76029-1
About Fujita Health University
Fujita Health University is a private university situated in Toyoake, Aichi, Japan. It was founded in 1964 and houses one of the largest teaching university hospitals in Japan in terms of the number of beds. With over 900 faculty members, the university is committed to providing various academic opportunities to students internationally. Fujita Health University has been ranked eighth among all universities and second among all private universities in Japan in the 2020 Times Higher Education (THE) World University Rankings. THE University Impact Rankings 2019 visualized university initiatives for sustainable development goals (SDGs). For the “good health and well-being” SDG, Fujita Health University was ranked second among all universities and number one among private universities in Japan. The university became the first Japanese university to host "THE Asia Universities Summit" in June 2021. The university’s founding philosophy is “Our creativity for the people (DOKUSOU-ICHIRI),” which reflects the belief that, as with the university’s alumni and alumnae, current students also unlock their future by leveraging their creativity.
Website: https://www.fujita-hu.ac.jp/en/index.html
About Associate Professor Katsuyuki Kunida from Fujita Health University
Dr. Katsuyuki Kunida is an Associate Professor at the School of Medicine at Fujita Health University, Japan. He obtained his Ph.D. in Medicine from Kyoto University, and his research focuses on system-theoretical understanding, prediction, and control of cell fate decision-making, cell motility, and metabolism, particularly within regenerative medicine and the prediction of pathological conditions. Dr. Kunida aims to advance preventive medicine through research focused on the precise prediction and control of cell systems. He has received multiple research awards and has over 20 publications to his credit.
About Mr. Tomoki Ohkubo from Nara Institute of Science and Technology
Mr. Tomoki Ohkubo is a PhD candidate in Computational Biology at Nara Institute of Science and Technology (NAIST). He received his B.S. in Precision Engineering and M.S. in Bioengineering from the University of Tokyo. He is also a biomedical engineer at Shimadzu Corporation, where he develops cell culture systems using microfluidics.
Funding information
This study was supported by the Next Generation Interdisciplinary Research Project of the Nara Institute of Science and Technology (NAIST) and AMED under Grant Numbers JP23wm0425017 and JP23tm0524001.
END