Using 3-D time-lapse movies to track the fate of misfolded proteins in yeast cells, the researchers determined that about 90 percent of aggregates form on the surface of the endoplasmic reticulum (ER), a location of protein synthesis in the cell. It had been thought that misfolded proteins spontaneously clump together in the cytosol, the fluid component of a cell's interior.
"Our findings have challenged the notion of the aggregation process as a passive consequence of accumulating misfolded proteins," says Stowers Investigator Rong Li, Ph.D., who led the study. Using budding yeast Saccharomyces cerevisae, a frequently used laboratory model in aging research, Stowers scientists experimentally used heat and other forms of stress to induce misfolded proteins to clump together.
Li and collaborators also found that the aggregation of misfolded proteins on the ER surface depends on the active synthesis of proteins by ribosomes. These molecular machines translate the cell's recipes for proteins. Guided by the recipe, the ribosome generates a linear polypeptide chain, the initial form of a protein.
The newly synthesized polypeptide folds into a distinctive three-dimensional structure resulting in a protein with a functional shape. Proteins that fail to fold correctly cannot perform their biological functions and are potentially toxic to cells. Thus, the aggregation of misfolded or unfolded proteins may help protect the cell and prevent their transfer to daughter cells during cell division, said Chuankai Zhou, a predoctoral researcher in the Li lab and first author of the paper.
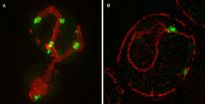
In addition to determining that protein aggregation is regulated and requires active translation, Stowers scientists revealed that the mitochondria, the cell's powerhouses, play a key role in the mobility of these protein aggregates. "We found the majority of aggregates on the surface of ER were in regions where ER and mitochondria come together, which is surprising but fits well with the view of regulated aggregation," says Zhou.
The current study builds upon previous research, published by the Li lab in 2011 in Cell, that revealed that most aggregates of unfolded proteins are retained by the mother yeast cell during the asymmetric cell division that characterizes this organism as well as stem cells. Budding yeast reproduce when only a small growth out of the mother yeast cell, a bud, becomes a daughter cell.
In the current paper, the scientists identified the quality control mechanism that limits the spreading of the misfolded protein aggregates to the bud and thereby the daughter cell. During the mitosis stage of budding yeast's division, aggregates of abnormal protein are tethered to well-anchored mitochondria in the mother cell. As a result, the mitochondria acquired by the bud are largely free of the abnormal aggregates.
"Remarkably, the majority of aggregates present in the mother cell did not follow the mitochondria that entered the bud, but they either underwent dissolution or remained associated with the mitochondria that were kept in the mother cell and only exhibited confined local mobility," describes Li. "As such, the bud-inherited mitochondria were largely devoid of aggregates."
Disrupting the aggregate-mitochondria association increases the mobility and leakage of mother-accumulated aggregates into the bud. Li and her colleagues also determined that the association of aggregates with mitochondria gradually declines in the mother budding yeast cells with advanced replicative age, likely contributing to their diminished ability to rejuvenate through asymmetric cell division.
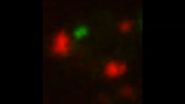
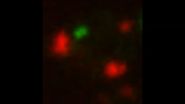
The researchers were able to visualize the process because the protein aggregates were labeled with different fluorescently tagged proteins that bind to aggregates. The Electron Microscopy and Imaging core centers at the Stowers Institute assisted with the research.
Other authors of the paper are Stowers researchers Brian D. Slaughter, Ph.D., Jay R. Unruh, Ph.D., Fengli Guo, Ph.D., Zulin Yu, Kristen Mickey, Akshay Narkar, Rhonda Trimble Ross, and Melainia McClain, Ph.D. Zhou, Mickey, Narkar, and Li also are affiliated with the University of Kansas Medical Center.
The study was supported by the Stowers Institute for Medical Research, the American Heart Association (predoctoral fellowship 13PRE16750001 to Zhou), and the National Institutes of Health (National Institute of General Medicine Sciences award RO1GM057063 to Li). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
VIDEO: Mitochondria dominate aggregate's motility. Red, mitochondria outer membrane; Green, aggregates.
Click here for more information.
Lay Summary of Findings
Patients with Parkinson's disease, cardiovascular disease, and cystic fibrosis may have something in common: cells in their disease-affected tissues may produce misfolded proteins that are incapable of functioning normally. In the current issue of the scientific journal Cell, Stowers Institute scientists report where the misfolded proteins clump together in a cell, and how the cell can prevent the passage of these defective molecules to its daughter cell. Stowers Investigator Rong Li, Ph.D., who headed the study, explains that during cell division, aggregates of these misfolded proteins normally are tethered to the mother cell. As a result, the daughter cell does not inherit the defective proteins that burden the mother cell. By identifying the quality control mechanisms that normally operate in cells, Li and other scientists are providing information that may prove relevant to treating disorders characterized by misfolded proteins.
INFORMATION:
About the Stowers Institute for Medical Research
The Stowers Institute for Medical Research is a non-profit, basic biomedical research organization dedicated to improving human health by studying the fundamental processes of life. Jim Stowers, founder of American Century Investments, and his wife, Virginia, opened the Institute in 2000. Since then, the Institute has spent over one billion dollars in pursuit of its mission.
Currently, the Institute is home to nearly 550 researchers and support personnel; over 20 independent research programs; and more than a dozen technology-development and core facilities.