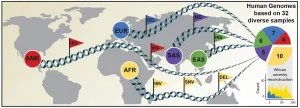
Building on this, scientists have carried out many sequencing projects over the last 20 years to identify and catalog genetic differences between an individual and the reference genome. Those differences usually focused on small single base changes and missed larger genetic alterations. Current technologies now are beginning to detect and characterize larger differences - called structural variants - such as insertions of several hundred letters. Structural variants are more likely than smaller genetic differences to interfere with gene function.
An international research team has now published an article in Science announcing a new, considerably more comprehensive reference dataset obtained using a combination of advanced sequencing and mapping technologies. The new reference dataset reflects 64 assembled human genomes, representing 25 different human populations from across the globe. Importantly, each of the genomes was assembled without guidance from the first human genome and as a result better captures genetic differences from different human populations. The study was led by scientists from the European Molecular Biology Laboratory Heidelberg (EMBL), the Heinrich Heine University Düsseldorf (HHU), The Jackson Laboratory for Genomic Medicine in Farmington, Conn. (JAX), and the University of Washington in Seattle (UW).
"With these new reference data, genetic differences can be studied with unprecedented accuracy against the background of global genetic variation, which facilitates the biomedical evaluation of genetic variants carried by an individual," emphasizes the co-first author of the study, Dr. Peter Ebert from the Institute of Medical Biometry and Bioinformatics at HHU. The distribution of genetic variants can differ substantially between population groups as a result of spontaneous and continuously occurring changes in the genetic material. If such a mutation is passed on over many generations, it can become a genetic variant specific to that population.
The new reference data provide an important basis for including the full spectrum of genetic variants in so-called genome-wide association studies. The aim is to estimate the individual risk of developing certain diseases such as cancer and to understand the underlying molecular mechanisms. This, in turn, can be used as a basis for more targeted therapies and preventative medicine.
This work might enable further applications in precision medicine. Drug efficacy, for example, can vary between individuals based on their genomes. The new reference data now represent the full range of different genetic variant types and incorporates human genomes of great diversity. Therefore, this new resource might contribute to developing novel approaches in personalized medicine, where the selection of therapies is tailored to a patient's individual genetic background.
This study builds on a new method published by these researchers last year in Nature Biotechnology) to accurately reconstruct the two components of a person's genome - one inherited from a person's father, one from a person's mother. When assembling a person's genome, this method eliminates the potential biases that could result from comparisons with an imperfect reference genome.
INFORMATION:
Source
Peter Ebert*, Peter A. Audano*, Qihui Zhu*, Bernardo Rodriguez-Martin*, David Porubsky, Marc Jan Bonder, Arvis Sulovari, Jana Ebler, Weichen Zhou, Rebecca Serra Mari, Feyza Yilmaz, Xuefang Zhao, PingHsun Hsieh, Joyce Lee, Sushant Kumar, Jiadong Lin, Tobias Rausch, Yu Chen, Jingwen Ren, Martin Santamarina, Wolfram Höps, Hufsah Ashraf, Nelson T. Chuang, Xiaofei Yang, Katherine M. Munson, Alexandra P. Lewis, Susan Fairley, Luke J. Tallon, Wayne E. Clarke, Anna O. Basile, Marta Byrska-Bishop, Andre Corvelo, Uday S. Evani, Tsung-Yu Lu, Mark J.P. Chaisson, Junjie Chen, Chong Li, Harrison Brand, Aaron M. Wenger, Maryam Ghareghani, William T. Harvey, Benjamin Raeder, Patrick Hasenfeld, Allison A. Regier, Haley J. Abel, Ira M. Hall, Paul Flicek, Oliver Stegle, Mark B. Gerstein, Jose M.C. Tubio, Zepeng Mu, Yang I. Li, Xinghua Shi, Alex R. Hastie, Kai Ye, Zechen Chong, Ashley D. Sanders, Michael C. Zody, Michael E. Talkowski, Ryan E. Mills, Scott E. Devine, Charles Lee#, Jan O. Korbel#, Tobias Marschall#, Evan E. Eichler#, Haplotype-resolved diverse human genomes and integrated analysis of structural variation, Science 2021
*Co-first authors #Co-senior and co-corresponding authors
Quotes
"For each human individual that participated in the study, we identified not one but two genomes - one for each set of chromosomes," says Jan Korbel, Ph.D., Head of Data Science at the European Molecular Biology Laboratory (EMBL) in Heidelberg who led the research at EMBL. Korbel added: "Humans have two sets of chromosomes which they receive from their parents. Previously we could not distinguish whether genetic variation came from one chromosome set or the other, and we were now able to solve this thanks to advances made by the Human Genome Structural Variation Consortium. It represents a remarkable achievement for the discovery of genetic variation in humans, which can now be studied much more comprehensively, leading the way to better find disease-causing genes."
"These genomes will pave the way to a new wave of scientific discoveries about the biology of the human genome and the connection between genetic variation and disease", says Bernardo Rodriguez-Martin, researcher at EMBL and co-first author. Rodriguez-Martin added: "As an example, we were able to estimate the age of highly mutagenic L1 repeats. Very surprisingly, although these sequences originated up to 3 Million of years ago, they continue to mutate the human genome frequently, which occasionally leads to diseases such as cancer."
"Just a few years ago, I would not have imagined that resolving genomes to this completeness would become possible so fast. This was enabled by exciting advances both of biotechnological and computational methods." says Dr. Peter Ebert, co-first author and computational biologist at Heinrich Heine University Düsseldorf, Germany. "Great to see this technology applied to a diversity panel of human genomes. These genome sequences will be an important resource for fundamental research and clinical genomics going forward."
Senior author Prof. Dr. Tobias Marschall, who led the research at HHU, added that "it was especially exciting to see that these new genome sequences enable a much more detailed analysis of data from standard sequencing technologies, which are routinely applied to millions of genomes by researchers and clinicians across the globe." He believes that "future studies to find associations between genetic variants and disease susceptibility will clearly benefit from this new approach."
"The first human genome sequence was a huge step forward, but was incomplete," said Charles Lee, Ph.D., FACMG, director and professor, The Jackson Laboratory for Genomic Medicine. "In addition to single base variation, we now know that structural variants also contribute very substantially to genomic differences between individuals. Our work provides a far more thorough and accurate window into that genomic variation across individuals and populations, and it represents an incredibly valuable new resource for the research community."
"Capturing the full spectrum of structural variation found in human genomes is vital for clinical applications," says Qihui Zhu, Ph.D., computational scientist. "These variants affect gene function and can contribute to diseases, drug response differences, and more. Knowing how they differ across individuals and across populations is needed to implement more effective genomic medicine."
"Each of these individual genomes is being resolved more completely for a fraction of the price of the first human genome" commented senior author, Evan Eichler, Professor of Genome Sciences, University of Washington School Medicine who was also a member of the original Human Genome Project. "We are discovering remarkable differences in genomic organization which have been missed until now, understanding these differences will enhance our ability to make genetic discoveries related to health and disease especially in groups that have been traditionally under-served by genomics research".
Peter Audano, co-first author, University of Washington School Medicine, adds, "the technology we have today can see into blindspots that have hidden information about diseases and our history. With these advances, we have discovered more than 100,000 structural variants, many of which are novel and affect genes or gene regulatory elements."
About EMBL
EMBL is Europe's flagship laboratory for the life sciences. Established in 1974 as an intergovernmental organisation, EMBL is supported by 27 member states, 2 prospective member states and 2 associate member states.
EMBL performs fundamental research in molecular biology, studying the story of life. The institute offers services to the scientific community; trains the next generation of scientists and strives to integrate the life sciences across Europe.
EMBL is international, innovative and interdisciplinary. Its more than 1800 staff, from over 80 countries, operate across six sites in Barcelona (Spain), Grenoble (France), Hamburg (Germany), Heidelberg (Germany), Hinxton (UK) and Rome (Italy). EMBL scientists work in independent groups and conduct research and offer services in all areas of molecular biology.
EMBL research drives the development of new technology and methods in the life sciences. The institute works to transfer this knowledge for the benefit of society.
https://www.embl.de/
About HHU
Heinrich Heine University Düsseldorf is one of the younger higher education institutions in the state of North Rhine-Westphalia - founded in 1965. Since 1988 our university has carried the name of one of the city's finest sons. Today around 35,000 students study at a modern campus under conditions ideally suited to academic life.
As a campus university where everything is close together, all buildings including the University Hospital and the specialised libraries are easily reachable. Our university departments enjoy an excellent reputation due to an exceptionally high number of collaborative research centres. Moreover, the state capital Düsseldorf provides an attractive environment with a high quality of life.
https://www.hhu.de/en/
About The Jackson Laboratory
The Jackson Laboratory is an independent, nonprofit biomedical research institution with more than 2,300 employees. Headquartered in Bar Harbor, Maine, it has a National Cancer Institute-designated Cancer Center, a genomic medicine institute in Farmington, Conn., and facilities in Ellsworth and Augusta, Maine, in Sacramento, Calif., and in Beijing and Shanghai, China. Its mission is to discover precise genomic solutions for disease and empower the global biomedical community in the shared quest to improve human health. For more information, please visit http://www.jax.org.
About University of Washington School Medicine
http://www.uwmedicine.org/school-of-medicine