(Press-News.org) DENVER – Armed with a catalog of hundreds of thousands of DNA and RNA virus species in the world’s oceans, scientists are now zeroing in on the viruses most likely to combat climate change by helping trap carbon dioxide in seawater or, using similar techniques, different viruses that may prevent methane’s escape from thawing Arctic soil.
By combining genomic sequencing data with artificial intelligence analysis, researchers have identified ocean-based viruses and assessed their genomes to find that they “steal” genes from other microbes or cells that process carbon in the sea. Mapping microbial metabolism genes, including those for underwater carbon metabolism, revealed 340 known metabolic pathways throughout the global oceans. Of these, 128 were also found in the genomes of ocean viruses.
“I was shocked that the number was that high,” said Matthew Sullivan, professor of microbiology and director of the Center of Microbiome Science at The Ohio State University.
Having mined this massive trove of data via advances in computation, the team has now revealed which viruses have a role in carbon metabolism and are using this information in newly developed community metabolic models to help predict how using viruses to engineer the ocean microbiome toward better carbon capture would look.
“The modeling is about how viruses may dial up or dial down microbial activity in the system,” Sullivan said. “Community metabolic modeling is telling me the dream data point: which viruses are targeting the most important metabolic pathways, and that matters because it means they’re good levers to pull on.”
Sullivan presented the research today (Feb. 17, 2024) at the annual meeting of the American Association for the Advancement of Science in Denver.
Sullivan was the virus coordinator for the Tara Oceans Consortium, a three-year global study of the impact of climate change on the world’s oceans and the source of 35,000 water samples containing the microbial bounty. His lab focuses on phages, viruses that infect bacteria, and their potential to be scaled up in an engineering framework to manipulate marine microbes into converting carbon into the heaviest organic form that will sink to the ocean floor.
“Oceans soak up carbon, and that buffers us against climate change. CO2 is absorbed as a gas, and its conversion into organic carbon is dictated by microbes,” Sullivan said. “What we’re seeing now is that viruses target the most important reactions in these microbial community metabolisms. This means we can start investigating which viruses could be used to convert carbon toward the kind we want.
“In other words, can we strengthen this massive ocean buffer to be a carbon sink to buy time against climate change, as opposed to that carbon being released back into the atmosphere to accelerate it?”
In 2016, the Tara team determined that carbon sinking in the ocean was related to the presence of viruses. It is thought that viruses help sink carbon when virus-infected carbon-processing cells cluster into larger, sticky aggregates that drop to the ocean floor. The researchers developed AI-based analytics to identify from thousands of viruses which few are “VIP” viruses to culture in the lab and work with as model systems for ocean geoengineering.
This new community metabolic modeling, developed by collaborator Professor Damien Eveillard of the Tara Oceans Consortium, helps them understand what unintended consequences might be of such an approach. Sullivan’s lab is taking these oceanic lessons learned and applying them to using viruses to engineer microbiomes in human settings to aid recovery from spinal cord injury, improve outcomes for infants born to mothers with HIV, combat infection in burn wounds, and more.
“The conversation we’re having is, ‘How much of this is transferable?’” said Sullivan, also a professor of civil, environmental and geodetic engineering. “The overall goal is engineering microbiomes toward what we think is something useful.”
He also reported on early efforts to use phages as geoengineering tools in an entirely different ecosystem: the permafrost in northern Sweden, where microbes both change the climate and respond to climate change as the frozen soil thaws. Virginia Rich, associate professor of microbiology at Ohio State, is co-director of the National Science Foundation-funded EMERGE Biology Integration Institute based at Ohio State that organizes the microbiome science at the Sweden field site. Rich also co-led previous research that identified a lineage of single-cell organisms in the thawing permafrost soil as a significant producer of methane, a potent greenhouse gas.
Rich co-organized the AAAS session with Ruth Varner of the University of New Hampshire, who co-directs the EMERGE Institute, which is focusing on better understanding how microbiomes respond to permafrost thaw and the resulting climate interactions.
Sullivan’s talk was titled “From ecosystems biology to managing microbiomes with viruses,” and was presented at the session titled “Microbiome-Targeted Ecosystem Management: Small Players, Big Roles.”
The oceans work is supported by the National Science Foundation, the Gordon and Betty Moore Foundation and Tara Oceans, and, in addition to the NSF, the soils work has been funded by the Department of Energy and the Grantham Foundation.
#
Contact: Matthew Sullivan, Sullivan.948@osu.edu
Written by Emily Caldwell, Caldwell.151@osu.edu
END
Embargoed until 1:30 p.m. ET, Saturday Feb. 17, 2024
DENVER – Imageomics, a new field of science, has made stunning progress in the past year and is on the verge of major discoveries about life on Earth, according to one of the founders of the discipline.
Tanya Berger-Wolf, faculty director of the Translational Data Analytics Institute at The Ohio State University, outlined the state of imageomics in a presentation on Feb. 17, 2024, at the annual meeting of the American Association for the Advancement of Science.
“Imageomics ...
Tokyo, Japan – Researchers from Tokyo Metropolitan University crafted replica stone age tools and used them for a range of tasks to see how different activities create traces on the edge. They found that a combination of macroscopic and microscopic traces can tell us how stone edges were used. Their criteria help separate tools used for wood-felling from other activities. Dated stone edges may be used to identify when timber use began for early humans.
For prehistoric humans, improvements in woodworking technology were revolutionary. While Paleolithic (early stone age) artifacts point to the use of wood for simple tools such as spears ...
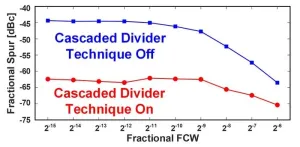
Two innovative design techniques lead to substantial improvements in performance in fractional-N phase locked loops (PLLs), report scientists from Tokyo Tech. The proposed methods are aimed to minimize unwanted signals known as fractional spurs, which typically plague PLLs used in many modern radar systems and wireless transceivers. These efforts could open doors to technological improvements in wireless communication, autonomous vehicles, surveillance, and tracking systems in beyond 5G era.
Many emerging and evolving technologies, such as self-driving vehicles, target tracking systems, and remote sensors, rely on the high-speed and error-free operation ...
DENVER — Young physician investigators interested in research careers in pulmonology, allergy and immunology, pediatric and related programs, are encouraged to submit basic science or clinical research abstracts by June 3, 2024, to be considered for participation in the 20th Annual Respiratory Disease Young Investigators’ Forum. This year’s Forum will take place October 17-20, 2024, in Denver.
The annual event provides career development and research opportunities for fellows and early career faculty. The Forum is a celebration of talent and ingenuity in respiratory medicine. Physician-scientists in fellowship ...
In an article published in the Journal of Pediatrics, researchers based in Brazil describe the case of a nine-year-old boy admitted to hospital with multiple symptoms and overlapping conditions that made diagnosis difficult, such as short stature, thin tooth enamel (dental enamel hypoplasia), moderate mental deficiency, speech delay, asthma, mildly altered blood sugar, and a history of recurring infections in infancy.
The team used exome sequencing, in which only the protein-coding portion of the genome is analyzed, to look for genetic mutations, and found them in GCK and BCL11B. ...
MIAMI, FLORIDA (Feb. 15, 2024) – Prominent oncologist and researcher Damian Green, M.D., will join Sylvester Comprehensive Cancer Center at the University Miami Miller School of Medicine this spring to lead its transplantation and cellular therapy services.
Green will serve as chief of Sylvester’s Division of Transplantation & Cellular Therapy, as well as assistant director of Translational Research, beginning March 1. He joins Sylvester from Fred Hutchinson Cancer Center in Seattle, where he built a distinguished track record in research and ...
By Wynne Parry
WOODS HOLE, Mass. – Like humans struggling to get through the COVID-19 pandemic, bacterial cells need social distancing to thwart viruses. But in some situations, such as inside elevators or within the candy-colored bacterial structures known as “pink berries,” staying apart just isn’t feasible.
Looking like spilled Nerds or Pop Rocks, the communal, multicellular pink berries litter the submerged surface of salt marshes in and around Woods Hole. New research conducted at the Marine Biological ...
Hospital-acquired acute kidney injury (HA-AKI) is a common complication in hospitalized patients that can lead to chronic kidney disease and is associated with longer hospital stays, higher health care costs and increased mortality. Given these negative consequences, preventing HA-AKI can improve hospitalized patient outcomes. However, anticipating HA-AKI onset is difficult due to a large number of contributing factors involved.
Researchers from Mass General Brigham Digital tested a commercial machine learning tool, the Epic Risk of HA-AKI predictive model, and found it was moderately successful at predicting risk of HA-AKI in recorded patient data. ...
(Toronto, February 16, 2024) JMIR Publications is pleased to announce that JMIR Bioinformatics and Biotechnology has passed the Scientific Quality Review by the US National Library of Medicine (NLM) for PubMed Central (PMC). This decision reflects the scientific and editorial quality of the journal. All articles published from 2022 onward will be found on PMC and PubMed after their technical evaluation.
Launched in 2020, JMIR Bioinformatics and Biotechnology is a sister journal of Journal of Medical Internet Research ...
Standard chemotherapy regimens, which are mostly based on testing in relatively young and healthy patients, may do more harm to older adults with cancer who often struggle with other health issues. New research, published yesterday in JAMA Network Open, shows that lowering the dose and adjusting the schedule of how chemotherapy is given to older adults with advanced cancer can make life better for patients, without compromising their treatment goals.
The study, which was led by researchers from the University of Rochester Medical Center’s Wilmot Cancer Institute, focuses on how well older people tolerate chemotherapy. More than 30 percent of patients benefitted from treatment regimen ...