New database to speed genetic discoveries
Tool lets any clinician contribute information about patients for analysis
2013-03-18
(Press-News.org) A new online database combining symptoms, family history and genetic sequencing information is speeding the search for diseases caused by a single rogue gene. As described in an article in the May issue of Human Mutation, the database, known as PhenoDB, enables any clinician to document cases of unusual genetic diseases for analysis by researchers at the Johns Hopkins University School of Medicine or the Baylor College of Medicine in Houston. If a review committee agrees that the patient may indeed have a previously unknown genetic disease, the patient and some of his or her family members may be offered free comprehensive genetic testing in an effort to identify the disease culprit.
"PhenoDB is much more useful than I even thought it would be," says Ada Hamosh, M.D., M.P.H., a professor in the McKusick-Nathans Institute of Genetic Medicine at the Johns Hopkins University School of Medicine. "Bringing all of this information together is crucial to figuring out what our genetic variations mean." The database is designed to capture a bevy of standardized information about phenotype, which Hamosh defines as "any characteristic of a person" — symptoms, personal and family health history, appearance, etc.
Hamosh and others developed PhenoDB for the Baylor-Hopkins Center for Mendelian Genomics (BHCMG), a four-year initiative that, together with its counterparts at Yale University and the University of Washington, is charged with uncovering the genetic roots of every disorder caused by a single faulty gene. There are an estimated 3,000 inherited disorders that have been described phenotypically in scientific papers but whose genetic causes have not yet been pinpointed, Hamosh says, but since many single-gene disorders are extremely rare, she suspects that many more have not yet made it into the literature.
The Centers for Mendelian Genomics have a powerful tool at their disposal, known as whole-exome sequencing. Just a few years ago, Hamosh explains, a geneticist trying to diagnose the cause of an inherited disease would have made an educated guess based on the patient's signs and symptoms about which gene might be at fault, and ordered a test of that gene. If the test came back negative for a mutation, she would order a test of a different gene, and so on. But whole-exome sequencing, in which about 90% of a person's genes are sequenced at one time, has been growing steadily cheaper, and it is this tool that the Centers will use to capture genetic sequencing information (whole-genome sequencing is the next step, but it remains too expensive for many uses, Hamosh notes, as it includes all of a person's DNA, most of which contains no genes).
However, making sense of the deluge of data yielded by whole-exome sequencing presents its own challenges. "The average person has tens of thousands of variations from the standard genetic sequence," Hamosh explains, "and we don't know what most of those variations mean." To parse these variations, she says, "one of the things that needs to change is that the lab doing the testing needs to have the whole phenotype, from head to toe." Researchers will then be better equipped to figure out which variations may or may not be relevant to a patient's illness. Another advantage of the database is that it enables colleagues at distant locations — such as Baylor and Johns Hopkins — to securely access the information and collaborate. Hamosh notes that the database enables different users to be afforded different levels of access — for example, a health provider will only be able to see the information he or she has entered — and that information is deidentified to protect patient privacy. In addition, providers must have patients' consent to be included in PhenoDB.
PhenoDB would be useful for any research project that seeks to match genomic information with its phenotypic effects, Hamosh says, and with that in mind, the Baylor-Hopkins Center for Mendelian Genomics has made the PhenoDB software available for free download at http://phenodb.net. She predicts that similar tools will soon be incorporated into electronic health records as well, so that "doctors will have patients' genomic information at their fingertips and can combine that with information about health history, disease symptoms and social situation to practice truly individualized medicine."
###
Other authors on the paper are Nara Sobreira, Julie Hoover-Fong, Corinne Boehm and David Valle, all of the Johns Hopkins University School of Medicine; V. Reid Sutton of Baylor College of Medicine; and François Schiettecatte of FS Consulting.
The Baylor-Hopkins Center for Mendelian Genomics is funded by the National Human Genome Research Institute (grant number 1U54HG006542).
Link to the paper: http://onlinelibrary.wiley.com/doi/10.1002/humu.22283/abstract END
ELSE PRESS RELEASES FROM THIS DATE:
2013-03-18
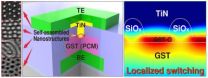
Daejeon, Republic of Korea, March 18, 2013 -- Nonvolatile memory that can store data even when not powered is currently used for portable electronics such as smart phones, tablets, and laptop computers. Flash memory is a dominant technology in this field, but its slow writing and erasing speed has led to extensive research into a next-generation nonvolatile memory called Phase-Change Random Access Memory (PRAM), as PRAM's operating speed is 1,000 times faster than that of flash memory.
PRAM uses reversible phase changes between the crystalline (low resistance) and amorphous ...
2013-03-18
Milan, 18 March 2013 – Markers such as CA9, CD31, CD34 and VEGFR1/2 in the primary tumours might serve as predictors of a good response to a sunitinib treatment in patients with metastatic clear cell renal cell carcinoma (ccRCC), according to a new study to be presented at the 28th Annual EAU Congress currently on-going in Milan.
"The inactivation of the von Hippel-Lindau gene (VHL) is a common event in ccRCC and finally leads to the induction of HIF1α target genes such as CA9 and VEGF," write the authors. "Besides VEGF, the VEGF and PDGF receptors also play an important ...
2013-03-18
Drs Lewis and Carré in the University's Department of Mechanical Engineering have been measuring the dynamic friction between the material of the ball and the skin on the fingertips and palm, and the mitts that some players choose to wear under different weather conditions. They're looking to answer one question: what's the best way to ensure that players don't fumble the ball?
"Catching and handling a ball with great skill and confidence is practically second nature to players at this level," says Dr Lewis. "But handling errors are still seen in professional rugby games ...
2013-03-18
CORAL GABLES, FL (March 18, 2012) -- University of Miami (UM) doctoral student in Environmental Science and Policy, David Shiffman was invited to tweet updates in real-time, at the International Congress of Conservation Biology, New Zealand, 2011. As a result, more than 100,000 twitter users worldwide saw at least one tweet from the conference, and nearly 200 people from more than 40 countries, on six continents shared at least one tweet from the conference -- greatly exceeding the number of conference attendees.
"While live-tweeting is not a new phenomenon, ...
2013-03-18
Marine biologists have, for the first time, found a whale skeleton on the ocean floor near Antarctica, giving new insights into life in the sea depths. The discovery was made almost a mile below the surface in an undersea crater and includes the find of at least nine new species of deep-sea organisms thriving on the bones.
The research, involving the University of Southampton, Natural History Museum, British Antarctic Survey, National Oceanography Centre (NOC) and Oxford University, is published today in Deep-Sea Research II: Topical Studies in Oceanography.
"The planet's ...
2013-03-18
The scientists looked at glaciers which behave independently from the ice sheet, despite having some physical connection to it, and those which are not connected at all.
The discovery, just published in Geophysical Research Letters, is important as it will help scientists improve the predictions of the future contribution of Greenland's ice to sea-level rise.
Using lasers which measure the height of the ice from space, and a recently completed inventory of Greenland's glaciers and ice caps, scientists from the European-funded ice2sea programme, were able to determine ...
2013-03-18
Washington, D.C.— It has long been believed that male lions are dependent on females when it comes to hunting. But new evidence suggests that male lions are, in fact, very successful hunters in their own right. A new report from a team including Carnegie's Scott Loarie and Greg Asner shows that male lions use dense savanna vegetation for ambush-style hunting in Africa. Their work is published in Animal Behavior.
Female lions have long been observed to rely on cooperative strategies to hunt their prey. While some studies demonstrated that male lions are as capable at hunting ...
2013-03-18
ANN ARBOR, Mich. — The long-term consequences of pneumonia can be more detrimental to a person's health than having a heart attack, according to joint research from the University of Michigan Health System and University of Washington School of Medicine.
Older adults who are hospitalized for pneumonia have a significantly higher risk of new problems that affect their ability to care for themselves, and the effects are comparable to those who survive a heart attack or stroke, according to the new findings in the American Journal of Medicine.
"Pneumonia is clearly not ...
2013-03-18
VIDEO:
This NASA research model, prepared on Mar. 15, 2013, from a space weather model known as ENLIL named after the Sumerian storm god, shows the way the CME was expected...
Click here for more information.
On March 17, 2013, at 1:28 a.m. EDT, the coronal mass ejection (CME) from March 15 passed by NASA's Advanced Composition Explorer (ACE) as it approached Earth. Upon interacting with the giant magnetic bubble surrounding Earth, the magnetosphere, the CME caused a kind of ...
2013-03-18
New research from an international team of scientists suggests evolution, or basic survival techniques adapted by early humans, influences the decisions gamblers make when placing bets.
The findings may help to explain why some treatment options for problem gamblers often don't work, the researchers say.
For the study, recently published in Frontiers in Psychology, scientists from McMaster University, the University of Lethbridge and Liverpool John Moores University examined how gamblers made decision after they won or lost.
They found that, like our ancestors, ...
LAST 30 PRESS RELEASES:
[Press-News.org] New database to speed genetic discoveries
Tool lets any clinician contribute information about patients for analysis