(Press-News.org) The COVID-19 pandemic has spurred genomic surveillance of viruses on an unprecedented scale, as scientists around the world use genome sequencing to track the spread of new variants of the SARS-CoV-2 virus. The rapid accumulation of viral genome sequences presents new opportunities for tracing global and local transmission dynamics, but analyzing so much genomic data is challenging.
"There are now more than a million genome sequences for SARS-CoV-2. No one had anticipated that number when we started sequencing this virus," said Russ Corbett-Detig, assistant professor of biomolecular engineering at UC Santa Cruz.
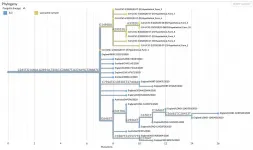
The sheer number of coronavirus genome sequences and their rapid accumulation makes it hard to place new sequences on a "family tree" showing how they are all related. But Corbett-Detig's group at the UC Santa Cruz Genomics Institute has developed a new method that does this with unprecedented speed. Called Ultrafast Sample Placement on Existing Trees (UShER), this powerful tool is described in a paper published May 10 in Nature Genetics.
UShER identifies the relationships between a user's newly sequenced viral genomes and all known SARS-CoV-2 virus genomes by adding them to an existing phylogenetic tree, a branching diagram like a family tree that shows how the virus has evolved in different lineages as it accumulates mutations.
"We are able to maintain a comprehensive phylogenetic tree of more than 1.2 million coronavirus sequences and update it with new sequences in real time. No other tool can handle trees of this size with a comparable efficiency," said first author Yatish Turakhia, a postdoctoral scholar at the Genomics Institute. "This helps us keep track of all variants in circulation, including new variants that are emerging."
This kind of sequence analysis can be used to discover new strains of the virus as they emerge and track their evolution and transmission dynamics. It can also be used to identify links between individual cases of coronavirus infection and to trace chains of transmission, an approach known as genomic contact tracing.
"The challenge is to get results soon enough to make meaningful predictions that public health agencies can use to try to control an outbreak," said Corbett-Detig, a corresponding author of the paper. "Our method is orders of magnitude faster than anything else out there, placing new samples in tenths of a second."
UShER and related data visualization tools are available to the research community through the UCSC SARS-CoV-2 Genome Browser, which also provides access to a wide range of data and results from ongoing scientific research on the virus, including new variants that are especially concerning.
"Our browser is the most comprehensive information resource for mutations appearing in the virus and what they mean for our battle against it," said coauthor David Haussler, professor of biomolecular engineering and director of the Genomics Institute. "Thanks to Russ's team, it includes the world's most comprehensive phylogenetic tree of the different lineages of the virus, and that tree continues to grow every week, as fast as new data appear."
Like all viruses, SARS-CoV-2 acquires mutations as it replicates and spreads. Most of these random variations in the genome sequence have no effect on the behavior of the virus, but researchers can still use them to identify different variants or strains of the virus, see how they are related, and determine if two samples are part of the same transmission chain.
Scientists have identified several important mutations that appear to make the virus more infectious. Variants of SARS-CoV-2 with these mutations are spreading more rapidly than other variants. Coauthor Angie Hinrichs, a UCSC Genome Browser engineer, used UShER to determine that one of these variants, known as B.1.1.7, entered the United States through several independent introductions. It is now the dominant strain in the United States.
Turakhia said he has begun using UShER to study a new variant that has emerged in India and appears to be spreading rapidly there. Known as B.1.617, this lineage of the virus has two mutations of potential concern to scientists. "We don't know yet how concerning it is, but it is important to track it," he said.
Viral genomics can reveal transmission chains not found through conventional contact tracing, Corbett-Detig said. This approach can help identify superspreader events, where one person transmitted the virus to many others, and it can also show that two cases from the same location are actually unrelated infections, not part of the same transmission chain, because the viral sequences differ too much.
"It's an approach that is likely to be valuable moving forward, so we're building the tools to enable people to do this in real time," he said. "If you want to know who transmitted the virus to whom, or where in the world a new sample may have come from, you need to take the samples from your community and project them onto the known phylogenetic tree of all the other SARS-CoV-2 genome sequences, and conventional phylogenetic methods just can't do this in a reasonable amount of time."
That's because conventional methods have to recalculate the entire tree every time new sequences are added, which is much too time-consuming when there are hundreds of thousands of sequences. UShER places samples onto an existing global phylogeny almost instantly, and it provides a local subtree of the added samples and their nearest neighbors so that their relationships can be visualized and examined in detail.
The researchers showed that UShER finds the right placement in 97% of cases. In the other 3%, incorrect placements are very close to the true site and still useful for contact tracing. UShER can also be used for quality control to quickly identify and remove low-quality sequences that may contain sequencing errors. The UShER results can be visualized and explored on the Nextstrain platform for interactive visualization of phylogenetic trees and maps of how the virus is spreading.
INFORMATION:
A training module for UShER is included in the CDC COVID-19 Genomic Epidemiology Toolkit (Module 3.3, including a video, slides, and links to more resources).
In addition to Turakhia, Corbett-Detig, Hinrichs, and Haussler, the coauthors of the paper include Bryan Thornlow and Landen Gozashti at UC Santa Cruz, Nicola De Maio at the European Bioinformatics Institute, and Robert Lanfear at Australian National University, Canberra. This work was funded by the Alfred P. Sloan Foundation and the National Institutes of Health.
PHILADELPHIA-- New research performed in mice models at Penn Medicine shows, mechanistically, how the infant lung regenerates cells after injury differently than the adult lung, with alveolar type 1 (AT1) cells reprograming into alveolar type 2 (AT2) cells (two very different lung alveolar epithelial cells), promoting cell regeneration, rather than AT2 cells differentiating into AT1 cells, which is the most widely accepted mechanism in the adult lung. These study findings, published today in Cell Stem Cell, show that the long-held assumption that AT1 ...
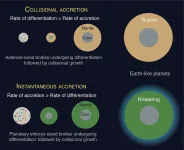
HOUSTON - (May 10, 2021) - The prospects for life on a given planet depend not only on where it forms but also how, according to Rice University scientists.
Planets like Earth that orbit within a solar system's Goldilocks zone, with conditions supporting liquid water and a rich atmosphere, are more likely to harbor life. As it turns out, how that planet came together also determines whether it captured and retained certain volatile elements and compounds, including nitrogen, carbon and water, that give rise to life.
In a study published in Nature Geoscience, Rice graduate student and lead author Damanveer Grewal and Professor Rajdeep Dasgupta show the competition between the time it takes for material to accrete into a protoplanet and the time the protoplanet ...
ITHACA, N.Y. - Voyager 1 - one of two sibling NASA spacecraft launched 44 years ago and now the most distant human-made object in space - still works and zooms toward infinity.
The craft has long since zipped past the edge of the solar system through the heliopause - the solar system's border with interstellar space - into the interstellar medium. Now, its instruments have detected the constant drone of interstellar gas (plasma waves), according to Cornell University-led research published in Nature Astronomy.
Examining data slowly sent back from more than 14 billion miles away, Stella Koch Ocker, a Cornell doctoral student in astronomy, has uncovered the emission. "It's very faint and monotone, because ...
Speech problems such as stammering or stuttering plague millions of people worldwide, including 3 million Americans. President Biden himself struggled with stuttering as a child and has largely overcome it with speech therapy. The cause of stuttering has long been a mystery, but researchers at Tufts University are beginning to unlock its causes and a strategy to develop potential treatments using a very curious model system - songbirds. In a study published today in Current Biology, the researchers were able to observe that a simple, reversible pharmacological treatment in zebra finches can stimulate rapid firing in a part of the brain that leads ...
Numerous disease development processes are linked to epigenetic modulation. One protein involved in the process of modulation and identified as an important cancer marker is BRD4. A recent study by the research group of Giulio Superti-Furga, Principal Investigator and Scientific Director at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, now shows that the supply of purines as well as the purine synthesis of a cell can influence BRD4 activity and thus play a role in the carcinogenesis process. The findings were published in Nature Metabolism.
Chromatin is a ...
New Curtin University research has found a bias among scientists toward colourful and visually striking plants, means they are more likely to be chosen for scientific study and benefit from subsequent conservation efforts, regardless of their ecological importance.
Co-author John Curtin Distinguished Professor Kingsley Dixon from Curtin's School of Molecular and Life Sciences was part of an international team that looked for evidence of an aesthetic bias among scientists by analysing 113 plant species found in global biodiversity hotspot the Southwestern Alps and mentioned in 280 research papers published between 1975 and 2020.
Professor Dixon said the study tested whether there was a relationship between research focus on plant species and characteristics ...
In cancer, a lot of biology goes awry: Genes mutate, molecular processes change dramatically, and cells proliferate uncontrollably to form entirely new tissues that we call tumors. Multiple things go wrong at different levels, and this complexity is partly what makes cancer so difficult to research and treat.
So it stands to reason that cancer researchers focus their attention where all cancers begin: the genome. If we can understand what happens at the level of DNA, then we can perhaps one day not just treat but even prevent cancers altogether.
This drive has led a ...
In the last few years, several technology companies including Google, Microsoft, and IBM, have massively invested in quantum computing systems based on microwave superconducting circuit platforms in an effort to scale them up from small research-oriented systems to commercialized computing platforms. But fulfilling the potential of quantum computers requires a significant increase in the number of qubits, the building blocks of quantum computers, which can store and manipulate quantum information.
But quantum signals can be contaminated by thermal noise generated by the movement of electrons. To prevent this, superconducting quantum systems must operate at ultra-low temperatures - less ...
NEW YORK, NY (May 10, 2021)--Scientists have discovered that many esophageal cancers turn on ancient viral DNA that was embedded in our genome hundreds of millions of years ago.
"It was surprising," says Adam Bass, MD, the Herbert and Florence Irving Professor of Medicine at Columbia University Vagelos College of Physicians and Surgeons and Herbert Irving Comprehensive Cancer Center, who led the study published May 10 in Nature Genetics.
"We weren't specifically searching for the viral elements, but the finding opens up a huge new array of potential cancer targets that I think will be extremely exciting as ways to enhance immunotherapy."
Fossil ...
DALLAS - May 10, 2021 - Scientists at UT Southwestern have discovered a key protein that helps the bacteria that causes Legionnaires' disease to set up house in the cells of humans and other hosts. The findings, published in Science, could offer insights into how other bacteria are able to survive inside cells, knowledge that could lead to new treatments for a wide variety of infections.
"Many infectious bacteria, from listeria to chlamydia to salmonella, use systems that allow them to dwell within their host's cells," says study leader Vincent Tagliabracci, Ph.D., assistant professor of molecular biology at UTSW and member of the Harold C. Simmons Comprehensive Cancer Center. "Better understanding the tools they use to make this happen is teaching us some interesting biochemistry and ...