(Press-News.org) Synthetic biologists have become increasingly creative in engineering yeast or bacteria to churn out useful chemicals — from fuels to fabrics and drugs — beyond the normal repertoire of microbes.
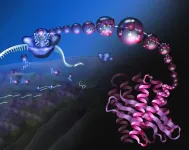
But a multi-university group of chemists has a more ambitious goal: to retool the cell's polypeptide manufacturing plants — the ribosomes that spin amino acids into protein — to generate polymer chains that are more elaborate than what can now be made in a cell or a test tube.
The $20 million research enterprise centered at the University of California, Berkeley, is now reporting significant progress toward that goal, as evidenced by three new papers that tackle three major hurdles: how to reprogram cells to supply the ribosome with building blocks other than the alpha-amino acids that make up all proteins today; how to predict which building blocks make the best substrates; and how to tweak the ribosome to incorporate these novel building blocks into polymers.
The ultimate goal of the National Science Foundation Center for Genetically Encoded Materials (C-GEM) is to make the translation system fully programmable, so that introducing mRNA instructions into the cell along with new building blocks — not the alpha-amino acids found today — will allow the ribosome to produce an unlimited variety of new molecular chains. These chains could form the basis for new bio-materials, new enzymes, even new drugs.
The papers, appearing in the journals Nature Chemistry and ACS Central Science, are the beginning of a playbook for reengineering the cellular synthetic machinery to produce never-before-seen polymers, including bio-polymers and circular polymers, which are called peptide macrocycles, with predetermined or completely unforeseen applications.
"C-GEM is working to biosynthesize molecules that have never before been made in a cell and that are designed to have unique properties. The tools could be applied broadly by polymer chemists, medicinal chemists and biomaterials scientists to generate bespoke materials with new functions," said C-GEM director Alanna Schepartz, the T.Z. and Irmgard Chu Distinguished Chair in Chemistry and professor of molecular and cell biology at UC Berkeley. "The ultimate goal is to expand the function and versatility of proteins and polypeptides, as both materials and pharmaceuticals."
One example, she said, would be to program the ribosome to synthesize a polymer that is a cross between spider silk — one of the toughest natural proteins — and nylon, a polymer now made in chemical reaction chambers. While spider silk can now be made in genetically engineered microbes, the technology C-GEM is developing could allow similar microbes to make an infinite variety of polymers blending the building blocks of silk and nylon, all of them new to chemists and with unique properties. The technology could also be used to make protein-like polymers more resistant to heat than natural proteins.
A powerful aspect of a programmable ribosome machine that can synthesize polymers is that it allows researchers to evolve the polymers to perfect their activity, just as proteins have evolved over hundreds of millions of years to improve the fitness of cells and organisms.
"We've had protein polymers evolving on the planet for billions of years, but we've been restricted in what those polymers are because the building blocks are the same 20 amino acids," said Jamie Cate, UC Berkeley professor of chemistry and of molecular and cell biology. "If we can develop a system where you can actually apply evolution to these new polymers, then it's like a platform that anybody who has a creative idea can use to evolve a polymer to something they want."
Such a system builds on the directed evolution of protein enzymes for which Frances Arnold, a UC Berkeley alumna, received the 2018 Nobel Prize in Chemistry.
"It's a step beyond what Frances Arnold did in developing directed evolution," Cate said. "She developed directed evolution for proteins. What we're trying to do is set up a way that you could do this for polymers never before evolved in nature."
Engineering an entirely new ribosome
In all cells, proteins are assembled by a nanomachine, the ribosome, that accepts instructions from an RNA molecule called messenger RNA (mRNA) — mRNA is akin to a working copy of a gene's DNA code — and reads those instructions to assemble a protein, amino acid by amino acid. Amazingly, the linear protein chain almost always folds into a well-defined 3D structure, ready to serve its evolved purpose: as an enzyme to catalyze reactions in the cell, as a structural component of the cell, or as a regulator of other cellular activities.
Ten years ago, retooling this complex nanomachine seemed impossible. But Schepartz's tenacity in pushing for a project to accomplish this goal resulted in C-GEM, which is three years into its first five-year funding cycle.
One of the center's goals is to supply the ribosome with building blocks — so-called monomers — other than alpha-amino acids. To achieve this goal, the C-GEM team focused on the enzymes that load amino acid monomers onto transfer RNA (tRNA), the molecules that ferry amino acids to the ribosome. Each tRNA is bar-coded to indicate which of the 20 amino acids it carries.
As reported in a Nature Chemistry paper published June 1 and co-authored by Schepartz and graduate students Riley Fricke and Cameron Swenson, the team discovered that one family of tRNA synthetases could load tRNA with four different non-alpha-amino acids. One of these was a building block of various polyketide therapeutics, including the antibiotics erythromycin and tetracycline.
"We identified enzymes that load tRNAs with monomers that differ structurally from anything that has been loaded on a tRNA before," Schepartz said. "One of the monomers is a precursor that could be used to assemble polyketide-like molecules. Scientists have been trying for decades to reengineer polyketide synthase modules to generate libraries of natural products. These studies have taught us a ton about the sophistication of these modules, but the engineering part has been very hard."
The novel monomers were willingly accepted by the native ribosome in the bacteria E. coli, demonstrating that it's possible to incorporate different types of chemistries into the normally all-amino acid protein polymer.
"Antibiotic resistance is an enormous problem," she added. "If we could help solve that problem by generating novel molecules whose functions encode unique modes of action, that would be an enormous contribution."
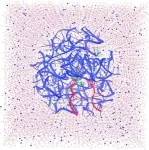
In a second paper, which appeared May 30 in ACS Central Science, lead author and postdoctoral fellow Chandrima Mujumdar, along with Cate and Schepartz, used cryogenic electron microscopy (cryo-EM) to obtain detailed structures of three related monomers — none of them alpha-amino acids — bound to the E. coli ribosome. The details show how these monomers bind — though much more poorly than do amino acids — and provide hints on how to alter the monomers or the ribosome to improve the ribosome's ability to use them to build novel polymers.
In a third paper, which appeared June 12 in Nature Chemistry, Cate, Schepartz and lead author Zoe Watson, a postdoctoral fellow, report the cryo-EM structure of the E. coli ribosome while binding normal alpha-amino acids. For this paper, the team collaborated with the company Schrödinger Inc. of San Diego, which uses computers to model protein binding. Ara Abramyan of Schrodinger used the cryo-EM structure as a starting point to run metadynamic simulations to help understand which non-natural monomers will react in the ribosome's catalytic center — the peptidyl transferase center (PTC) — and which will not.
Schepartz and Cate emphasized that all of these tweaks to the ribosomal system must work inside a living cell independently of the normal ribosomes so that the production of new polymers does not interfere with the day-to-day protein production necessary for life.
"We want enzymes — synthetases — and ribosomes that could be used in a cell, because that's how this work will be scalable," Schepartz said. "That goal requires robust ribosomes, great enzymes and a lot of understanding about the chemistry of how these complex molecular machines work. It’s a hard problem, but lots of fun. And we get to expose students and postdocs to some really great science."
Among the authors are C-GEM investigators Matthew Francis of UC Berkeley, Scott Miller of Yale University, Abhishek Chatterjee of Boston College, Bhavana Shah and Zhonqi Zhang of Amgen Inc., and C-GEM managing director Sarah Smaga. Schepartz is also a member of the Chan Zuckerberg Biohub and the California Institute for Quantitative Biosciences (QB3). Cate is a member of the Innovative Genomics Institute. Both Schepartz and Cate are faculty scientists at Lawrence Berkeley National Laboratory.
NSF provided most of the funding under grant CHE 2002182.
END
Retooling the ribosomal translation machine could expand chemical repertoire of cells
New research shows progress on playbook for programming ribosome to make diverse polymers
2023-06-13
ELSE PRESS RELEASES FROM THIS DATE:
Sickle cell disease is 11 times more deadly than previously recorded
2023-06-13
A new analysis provides a more complete picture of sickle cell disease mortality burden by combining disease prevalence data in different age groups and trends in overall survival when factoring in resulting secondary conditions.
When looking across all deaths, sickle cell disease is a leading cause of mortality in children under 5 years as well as in youth 5–14 years and adults 15–49 years.
Half a million babies were born with sickle cell disease in 2021, and 79% of these infants were in sub-Saharan Africa.
The largest increases ...
New approaches to evaluating water interventions around the globe
2023-06-13
Billions of people around the world face water insecurity. Although there are numerous projects from governments, NGOs, and private corporations who are committed to providing safely managed water and sanitation by 2030, a new study advocates for more holistic evaluation of water, sanitation, and hygiene (WASH) interventions.
According to the study by Justin Stoler, associate professor in the University of Miami College of Arts and Sciences Department of Geography and Sustainable Development, issues ...
ASCAP introduces slate of AI initiatives to help music creators navigate the future while protecting their work
2023-06-13
NEW YORK, June 13, 2023 – With the potential for artificial intelligence (AI) to create both massive disruption and great opportunity within the music industry, ASCAP — the only US PRO that operates on a not-for-profit basis — is introducing a slate of AI initiatives to help music creators navigate the future while protecting their work. These newly announced initiatives include: adoption by the ASCAP Board of Directors of a set of key ASCAP AI principles, creator education, startup incubation and policy development. Building upon ASCAP’s strong ...
Study in mice links heat-damaged DNA in food to possible genetic risks
2023-06-13
Researchers have newly discovered a surprising and potentially significant reason why eating foods frequently cooked at high temperatures, such as red meat and deep-fried fare, elevates cancer risk. The alleged culprit: DNA within the food that’s been damaged by the cooking process.
As shown for the first time known to the authors, this study by Stanford scientists and their collaborators at the National Institute of Standards and Technology (NIST), the University of Maryland, and Colorado State University reveals that components of heat-marred DNA can be absorbed during digestion and incorporated into the DNA of the consumer. That uptake directly ...
UC Irvine neuroscientists develop ‘meta-cell’ to move Alzheimer’s fight forward
2023-06-13
Irvine, Calif., June 13, 2023 – University of California, Irvine neuroscientists probing the gene changes behind Alzheimer’s disease have developed a process of making a “meta-cell” that overcomes the challenges of studying a single cell. Their technique has already revealed important new information and can be used to study other diseases throughout the body. Details about the meta-cell – created by researchers with the UCI Institute for Memory Impairments and Neurological Disorders, known as UCI MIND – were published in the online journal Cell Press.
Technologies called transcriptomics that study sets of RNA within organisms ...
What’s an underrated way to study decisions? Think out loud
2023-06-13
Think fast: if you’re running a race and overtake the person in second place, what position are you in? Many people instinctively respond that you’re in first place. However, upon reflection, some people realize the correct answer is that you’re now in second place: the former number-two runner slipped into third as you overtook them.
Trick questions of this kind are invaluable to cognitive scientists because they shed light on the cognitive quirks that shape our decision-making. “These aren’t just trick questions,” explained Nick Byrd, a philosopher-scientist and Intelligence Community Fellow who led the research at Stevens ...
Pass the salt: This space rock holds clues as to how Earth got its water
2023-06-13
Sodium chloride, better known as table salt, isn't exactly the type of mineral that captures the imagination of scientists. However, a smattering of tiny salt crystals discovered in a sample from an asteroid has researchers at the University of Arizona Lunar and Planetary Laboratory excited, because these crystals can only have formed in the presence of liquid water.
Even more intriguing, according to the research team, is the fact that the sample comes from an S-type asteroid, a category known to mostly lack hydrated, or water-bearing, minerals. The discovery strongly suggests ...
Understanding tumor microenvironment with photoacoustic spectral analysis
2023-06-13
Tumors are not just isolated clumps of abnormal cells but are associated with more complex system known as the “tumor microenvironment” (TME). Over the past few years, research has revealed that the TME consists of a complex combination of blood vessels, connective tissue, and a matrix of extracellular proteins and molecules. Most importantly, the composition of the TME is different from that of nearby healthy tissues. In particular, the lipid and collagen contents of tumors differ from those of normal tissues, making them important potential biomarkers for diagnosing various types of ...
Special issue of JICM on global public health and the Declaration of Astana
2023-06-13
A special focus issue of the peer-reviewed Journal of Integrative and Complementary Medicine (JICM) on Global Public Health is a response to the Declaration of Astana, developed by the World Health Organization and UNICEF. With contributions from traditional, complementary, and integrative medicine (TCIM) scholars and thought leaders around the world, the issue showcases research and scholarship examining the opportunities and challenges that TCIM approaches offer global health to governments and health service providers seeking to fulfill ...
ASU establishes ʻĀkoʻakoʻa, a new collaborative effort to seed renewed connection between human and coral communities in Hawaiʻi
2023-06-13
With a group of core partners, Arizona State University is creating a new $25 million collaboration to preserve and restore vitality to Hawaiʻi’s coral reefs and the health of its coastlines.
The community-based effort looks to fuse state-of-the-art science programs with the leadership and cultural knowledge of Hawaiʻi’s community partners to enable coastal and reef sustainability for generations to come. Named ʻĀkoʻakoʻa (pronounced ah kō-a kō-a), the effort shares a dual meaning ʻto assemble’ and ‘coral.’
“For ...
LAST 30 PRESS RELEASES:
Scientists develop new gut health measure that tracks disease
Rice gene discovery could cut fertiliser use while protecting yields
Jumping ‘DNA parasites’ linked to early stages of tumour formation
Ultra-sensitive CAR T cells provide potential strategy to treat solid tumors
Early Neanderthal-Human interbreeding was strongly sex biased
North American bird declines are widespread and accelerating in agricultural hotspots
Researchers recommend strategies for improved genetic privacy legislation
How birds achieve sweet success
More sensitive cell therapy may be a HIT against solid cancers
Scientists map how aging reshapes cells across the entire mammalian body
Hotspots of accelerated bird decline linked to agricultural activity
How ancient attraction shaped the human genome
NJIT faculty named Senior Members of the National Academy of Inventors
App aids substance use recovery in vulnerable populations
College students nationwide received lifesaving education on sudden cardiac death
Oak Ridge National Laboratory launches the Next-Generation Data Centers Institute
Improved short-term sea level change predictions with better AI training
UAlbany researchers develop new laser technique to test mRNA-based therapeutics
New water-treatment system removes nitrogen, phosphorus from farm tile drainage
Major Canadian study finds strong link between cannabis, anxiety and depression
New discovery of younger Ediacaran biota
Lymphovenous bypass: Potential surgical treatment for Alzheimer's disease?
When safety starts with a text message
CSIC develops an antibody that protects immune system cells in vitro from a dangerous hospital-acquired bacterium
New study challenges assumptions behind Africa’s Green Revolution efforts and calls for farmer-centered development models
Immune cells link lactation to long-lasting health
Evolution: Ancient mosquitoes developed a taste for early hominins
Pickleball players’ reported use of protective eyewear
Changes in organ donation after circulatory death in the US
Fertility preservation in people with cancer
[Press-News.org] Retooling the ribosomal translation machine could expand chemical repertoire of cellsNew research shows progress on playbook for programming ribosome to make diverse polymers