(Press-News.org) Microbes are part of the food we eat and can influence our own microbiome, but we know very little about the microbes in our foods. Now, researchers have developed a database of the “food microbiome” by sequencing the metagenomes of 2,533 different foods. They identified 10,899 food-associated microbes, half of which were previously unknown species, and showed that food-associated microbes account for around 3% of the adult and 56% of the infant gut microbiome on average. The study published August 29 in the journal Cell, and the database is available as an open access resource.
“This is the largest survey of microbes in food,” says co-senior author and computational microbiologist Nicola Segata (@nsegata) of the University of Trento and the European Institute of Oncology in Milan. “We can now start to use this reference to better understand how the quality, conservation, safety, and other characteristics of food are linked with the microbes they contain.”
Traditionally, microbes in food have been studied by culturing them one-by-one in the lab, but this process is slow and time consuming, and not all microbes can be easily cultured. To characterize the food microbiome more comprehensively and efficiently, the researchers leveraged metagenomics, a molecular tool that enabled them to simultaneously sequence all the genetic material within each food sample. Metagenomics is often used to characterize the human microbiome or analyze environmental samples but hasn’t previously been used to investigate food at a large scale.
“Food microbiologists have been studying foods and testing for food safety for well over a hundred years now, but we’ve underutilized modern DNA sequencing technologies,” says co-senior author and microbiologist Paul Cotter (@pauldcotter) of Teagasc, APC Microbiome Ireland and VistaMilk Ireland. “This is the starting point for a new wave of studies in the field where we make full use of the molecular technology available.”
Altogether, the team analyzed 2,533 food-associated metagenomes from 50 countries, including 1,950 newly sequenced metagenomes. These metagenomes came from a variety of food types, of which 65% were dairy sources, 17% were fermented beverages, and 5% were fermented meats.
These metagenomes comprised genetic material from 10,899 food-associated microbes categorized into 1,036 bacterial and 108 fungal species. Similar foods tended to harbor similar types of microbes—for example, the microbial communities in different fermented beverages were more similar to each other than to the microbes in fermented meat—but there was more variation between dairy products, likely due to the larger number of dairy products surveyed.
Though the researchers didn’t identify many overtly pathogenic bacteria in the food samples, they did identify some microbes that might be less desirable due to their impact on food flavor or preservation. Knowing which microbes “belong” in different types of food could help producers—both industrial and small-scale—to produce more consistent and desirable products. It could also help food regulators define which microbes should and should not be in certain types of food and to authenticate the identity and origins of “local” foods.
“One thing that was striking is that some microbes are present and performing similar functions in even quite different foods, and at the same time, we showed that foods in each local facility or farm have unique characteristics,” says Segata. “This is important because it could further improve the idea of the specificity and the quality of local foods, and we could even use metagenomics to authenticate foods coming from a given facility or location.”
Understanding the food microbiome could also have implications for human health as some of the microbes we eat could become stable members of our own microbiomes. To examine overlaps between food-associated microbes and the human microbiome, the team compared their new database with 19,833 previously sequenced human metagenomes. They showed that food-associated microbial species compose around 3% of the gut microbiome of adults and more than 50% of the gut microbiomes of newborns.
“This suggests that some of our gut microbes may be acquired directly from food, or that historically, human populations got these microbes from food and then those microbes adapted to become part of the human microbiome,” says Segata. “It might seem like only a small percentage, but that 3% can be extremely relevant for their function within our body. With this database, we can start surveying at a large scale how the microbial properties of food could impact our health.”
The study was one of the main outputs from the MASTER EU consortium, an EU-funded initiative spanning 29 partners across 14 countries that aims to characterize the presence and function of microbes throughout the entire food chain.
“In the future, we want to explore the diversity of these food microbiomes with respect to different foods, cultures, lifestyles, and populations,” says Cotter.
###
This research was supported by Horizon 2020 of Horizon Europe, the Italian Ministry of Foreign Affairs and International Cooperation, the European Research Council, the National Cancer Institute of the National Institutes of Health, the Spanish Ministry of Science and Innovation, the Science Foundation Ireland, and the Irish Department of Agriculture, Food and the Marine.
Cell, Carlino et al., “Unexplored microbial diversity from 2,500 food metagenomes and links with the human microbiome” https://cell.com/cell/fulltext/S0092-8674(24)00833-X
Cell (@CellCellPress), the flagship journal of Cell Press, is a bimonthly journal that publishes findings of unusual significance in any area of experimental biology, including but not limited to cell biology, molecular biology, neuroscience, immunology, virology and microbiology, cancer, human genetics, systems biology, signaling, and disease mechanisms and therapeutics. Visit http://www.cell.com/cell. To receive Cell Press media alerts, contact press@cell.com.
END
Researchers at Queen Mary University of London have made a groundbreaking discovery about how starfish manage to survive predatory attacks by shedding their own limbs. The team has identified a neurohormone responsible for triggering this remarkable feat of self-preservation.
Autotomy, the ability of an animal to detach a body part to evade predators, is a well-known survival strategy in the animal kingdom. While lizards shedding their tails are a familiar example, the mechanisms behind this process remain largely mysterious.
Now, scientists have unveiled a key piece of the puzzle. By studying the common European starfish, ...
About The Study: Postmenopausal women with historical hormone therapy (HT) use were biologically younger than those not receiving HT, with a more evident association observed in those with low socioeconomic status. The biological aging discrepancy mediated the association between HT and decreased mortality. Promoting HT in postmenopausal women could be important for healthy aging.
Corresponding Author: To contact the corresponding author, Chenglong Li, PhD, email chenglongli@bjmu.edu.cn.
To access the embargoed study: Visit our For The Media website at this link https://media.jamanetwork.com/
(doi:10.1001/jamanetworkopen.2024.30839)
Editor’s ...
About The Study: The findings of this study of women ages 18 or older diagnosed with stage I to III breast cancer between 2010 and 2018 suggest that residing in persistently impoverished neighborhoods is associated with poor tumor characteristics and increased mortality.
Corresponding Author: To contact the corresponding author, Samilia Obeng-Gyasi, MD, MPH, email samilia.obeng-gyasi@osumc.edu.
To access the embargoed study: Visit our For The Media website at this link https://media.jamanetwork.com/
(doi:10.1001/jamanetworkopen.2024.27755)
Editor’s Note: Please see the article for additional information, including other authors, ...
About The Study: Inhaler prescriptions filled by Centers for Medicare & Medicaid Services beneficiaries in 2022 resulted in an estimated 1.15 million metric tons of carbon dioxide equivalent emissions, equivalent to 226,960 homes’ yearly electricity use. Metered-dose inhalers were responsible for nearly all inhaler-related emissions, with the largest contribution arising from short-acting β-agonist medications. Although dry-powder and soft-mist inhalers had substantially lower emissions, they accounted for a disproportionate amount of spending, representing nearly two-thirds ...
How are rodents able to navigate pitch-black subway tunnels or other dark environments so adeptly, despite not being able to rely on vision?
With the assistance of a novel motion simulator, researchers at Bar-Ilan University in Israel have discovered that rats rely on airflow to navigate their surroundings. When they move, the flow of air relative to their bodies provides crucial information, complementary to their sense of balance, to perceive their own motion in space. This might explain their agility in the dark as they scurry through pipes and tunnels, ...
PHILADELPHIA – A new tool for monitoring immune health patterns over time has revealed how a pair of checkpoint inhibitor therapies works together to recruit new cancer-fighting T cells with every infusion. Findings from the use of the new tool, developed by researchers at the University of Pennsylvania Perelman School of Medicine and Penn Medicine’s Abramson Cancer Center (ACC), were published today in Cancer Cell. The study challenges fundamental assumptions about how a common immunotherapy ...
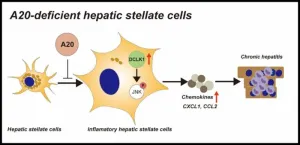
Researchers from Tokyo Medical and Dental University (TMDU) determine how a protein called A20 can regulate the inflammatory response to suppress chronic hepatitis
Tokyo, Japan – Many individuals worldwide suffer from chronic liver disease (CLD), which poses significant concerns for its tendency to lead to hepatocellular carcinoma or liver failure. CLD is characterized by inflammation and fibrosis. Certain liver cells, called hepatic stellate cells (HSCs), contribute to both these characteristics, but how they are specifically involved in the inflammatory response is not ...
The Microbiology Society, one of the largest microbiology societies in Europe, is pleased to announce a new three year Publish and Read offering with German consortium ZB Med – Information Centre for Life Sciences, available to its 400 member institutions and 2,000 hospitals. This agreement was established in partnership with HARRASSOWITZ, the Society’s representative agency in Germany.
From 2025, member institutions can join this consortium-wide Publish and Read agreement to enjoy discounted pricing. For participating institutions, the ...
Researchers at Nano Life Science Institute (WPI-NanoLSI), Kanazawa University, implement modifications to their high-speed atomic force microscopy that simultaneously improve resolution and speed, while enabling direct measurements of 3D structures to provide conclusive evidence of a contested hydration layer forming as calcite dissolves.
Understanding the dissolution processes of minerals can provide key insights into geochemical processes. Attempts to explain some of the observations during the dissolution of calcite (CaCO3) have led to the hypothesis that a hydration layer forms, although this has been contested. Hydration layers are also ...
A new study has found an answer for a long-lasting question in aging research - Is Alzheimer’s disease-dementia a form of accelerated aging or is there a different path that can lead us to healthier brain aging? In an international effort, the researchers mapped 1.65 million cells from 437 aging brains, and uncovered distinct paths of cellular change in the aging brains, with one leading to Alzheimer’s disease and the other to an alternative form brain aging. They also point to specific cell signatures predicted to advance disease once they appear in the aging ...