A new CRISPR-based gene-editing tool has been developed which could lead to better treatments for patients with genetic disorders. The tool is an enzyme, AsCas12f, which has been modified to offer the same effectiveness but at one-third the size of the Cas9 enzyme commonly used for gene editing. The compact size means that more of it can be packed into carrier viruses and delivered into living cells, making it more efficient. Researchers created a library of possible AsCas12f mutations and then combined selected ones to engineer an AsCas12f enzyme with 10 times more editing ability than the original unmutated type. This engineered AsCas12f has already been successfully tested in mice and has the potential to be used for new, more effective treatments for patients in the future.
By now you have probably heard of CRISPR, the gene-editing tool which enables researchers to replace and alter segments of DNA. Like genetic tailors, scientists have been experimenting with “snipping away” the genes that make mosquitoes malaria carriers, altering food crops to be more nutritious and delicious, and in recent years begun human trials to overcome some of the most challenging diseases and genetic disorders. The potential of CRISPR to improve our lives is so phenomenal that in 2020, researchers Jennifer Doudna and Emmanuelle Charpentier, who developed the most precise version of the tool named CRISPR-Cas9, were awarded the Nobel Prize in chemistry.
But even Cas9 has limitations. The common way to deliver genetic material into a host cell is to use a modified virus as a carrier. Adeno-associated viruses (AAVs) are not harmful to patients, can enter many different types of cells to introduce CRISPR enzymes like Cas9, and have a lower likelihood of provoking an undesired immune response compared to some other methods. However, like any parcel delivery service, there is a size limit. “Cas9 is at the very limit of this size restriction, so there has been a demand for a smaller Cas protein that can be efficiently packaged into AAV and serve as a genome-editing tool,” explained Professor Osamu Nureki from the Department of Biological Sciences at the University of Tokyo.
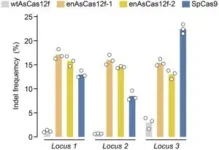
Its large size means that Cas9 can lack efficiency when used for gene therapy. So, a large multi-institutional team worked to develop a smaller Cas enzyme that is just as active, but more efficient. The researchers selected an enzyme called AsCas12f, from the bacteria Axidibacillus sulfuroxidans. The advantage of this enzyme is that it is one of the most compact Cas enzymes found to date and less than one-third the size of Cas9. However, in previous tests it showed barely any genome activity in human cells.
“Using a screening method called deep mutational scanning, we assembled a library of potential new candidates by substituting each amino acid residue of AsCas12f with all 20 types of amino acids on which all life is based. From this, we identified over 200 mutations that enhanced genome-editing activity,” explained Nureki. “Based on insights gained from the structural analysis of AsCas12f, we selected and combined these enhanced-activity amino acid mutations to create a modified AsCas12f. This engineered AsCas12f has more than 10 times the genome-editing activity compared to the usual AsCas12f type and is comparable to Cas9, while maintaining a much smaller size.”
The team has already carried out animal trials with the engineered AsCas12f system, partnering it with other genes and administering it to live mice. Administering treatments directly into the body is preferable to extracting cells, editing them in a lab and reinserting them into patients, which is more time-intensive and costly. The success of the tests showed that engineered AsCas12f has the potential to be used for human gene therapies, such as treating hemophilia, a disease in which the blood does not clot normally.
The team discovered numerous potentially effective combinations for engineering an improved AsCas12f gene-editing system, so the researchers acknowledge the possibility that the selected mutations may not have been the most optimal of all the available mixes. As a next step, computational modeling or machine learning could be used to sift through the combinations and predict which might offer even better improvements.
“Elevating AsCas12f to exhibit genome-editing activity comparable to that of Cas9 is a significant achievement and serves as a substantial step in the development of new, more compact genome-editing tools,” said Nureki. “For us the crucial aspect of gene therapy is its potential to genuinely help patients. Using the engineered AsCas12f we developed, our next challenge is to actually administer gene therapy to aid people suffering from genetic disorders.”
###
Paper Title:
Tomohiro Hino, Satoshi N. Omura, Ryoya Nakagawa, Tomoki Togashi, Satoru N. Takeda, Takafumi Hiramoto, Satoshi Tasaka, Hisato Hirano, Takeshi Tokuyama, Hideki Uosaki, Soh Ishiguro, Madina Kagieva, Hiroyuki Yamano, Yuki Ozaki, Daisuke Motooka, Hideto Mori, Yuhei Kirita, Yoshiaki Kise, Yuzuru Itoh, Satoaki Matoba, Hiroyuki Aburatani, Nozomu Yachie, Tautvydas Karvelis, Virginijus Siksnys, Tsukasa Ohmori, Atsushi Hoshino, Osamu Nureki. “An AsCas12f-based compact genome editing tool derived by deep mutational scanning and structural analysis” Cell. https://doi.org/10.1016/j.cell.2023.08.031
Funding:
This research was supported by the Research Council of Lithuania for the support of European Joint Programme on Rare Diseases project GET-READY (grant number S-EJPRD-21-1), Japan Foundation for Applied Enzymology, AMED Grant Numbers JP22fk0410037 and JP22ae0201007, AMED Grant Numbers JP233fa627001 and JP19am0401005, the Platform Project for Supporting Drug Discovery and Life Science Research from AMED under Grant Number JP22am0101115, and the Cabinet Office, Government of Japan, Public/Private R&D Investment Strategic Expansion Program (PRISM) Grant Number JPJ008000.
Useful Links :
Graduate School of Science: https://www.s.u-tokyo.ac.jp/en/
Department of Biological Sciences: http://www.bs.s.u-tokyo.ac.jp/english/
Declaration of interests:
T.H., S.N.O., R.N., S.M., T.O., A. H., O.N. have filed a patent application related to this work. The remaining authors declare no competing interests.
Research Contact:
Professor Osamu Nureki
Department of Biological Sciences
Graduate School of Science
The University of Tokyo
7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8654, Japan
Email: nureki@bs.s.u-tokyo.ac.jp
Tel.: 03-5841-4392
Press contact:
Mrs. Nicola Burghall
Public Relations Group, The University of Tokyo,
7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8654, Japan
press-releases.adm@gs.mail.u-tokyo.ac.jp
About the University of Tokyo
The University of Tokyo is Japan's leading university and one of the world's top research universities. The vast research output of some 6,000 researchers is published in the world's top journals across the arts and sciences. Our vibrant student body of around 15,000 undergraduate and 15,000 graduate students includes over 4,000 international students. Find out more at www.u-tokyo.ac.jp/en/ or follow us on Twitter at @UTokyo_News_en.
END